The abundance of dinitrogen (N2) in the earth's atmosphere and in other planetary atmospheres displays just how energetically stable the N≡N bond is. Having a bond enthalpy of nearly 950 kJ/mol ensures dinitrogen is the strongest existing bond, and keeps dinitrogen significantly nonreactive. Industrially, nitrogen is converted to ammonia in the Haber-Bosch process, requiring a catalyst, temperatures in excess of 200 C and at pressures in excess of 150 atm. Fertilizer from ammonia produced via the harsh conditions of the Haber-Bosch process accounts for over half the world's food production.
But yet, nature, utilizing nitrogenase, performs the very same reaction at ambient conditions. The prevalence of nitrogen atoms in proteins, DNA, RNA, and other bio-molecules displays its biological importance. Nitrogen is, clearly, one of the fundamental building blocks of life and, staggeringly, ALL of it is fixed, or made usable, by a single enzyme: nitrogenase. This fixed nitrogen is then incorporated into every single biological macromolecule. The stream of life truly begins here, at nitrogenase!
 |
| Fig. 1: The heterotetrameric structure of nitrogenase, pictured with homodimeric Fe protein subunits. (PDB code: 1N2C. Figure generated using Pymol.) |
Reaction
Overall Reaction: N2 + 16 ATP + 8 e- + 8 H+ ⇒ 2 NH3 + H2 + 16 ADP (ΔGrxn = - 93 kJ/mol)
Structure & Mechanism
Despite the continuing effort of numerous scientists, not much light has been shed on the mechanism of nitrogenase action. This is due, significantly, to the difficulty in trapping the highly reactive N-intermediates of the reaction. As a result, nitrogenase has been dubbed 'The Everest of Enzymes', displaying both the complexity of the mechanism and the enzyme's legendary status in the scientific community. To find out more, read paper 3 of the previous blog entry.
Yet, there is far more to be said about what is known of nitrogenase structure. To be concise, I will restrict myself to 3 key features of the enzyme: (1) Unique FeS clusters & cofactors, (2) electron transport in the enzyme, and finally (3) mechanism of preventing oxidation.
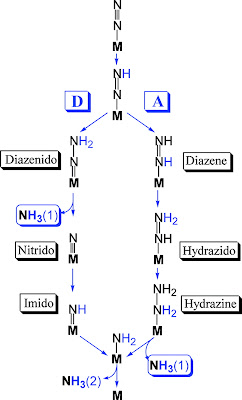
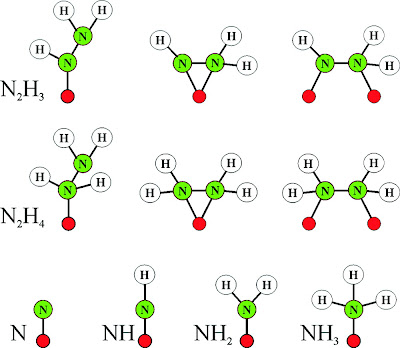
 |
| Fig. 2: Cofactor ensemble of nitrogenase(3) |
(1) Unique FeS Clusters: As mentioned earlier, nitrogenase contains 3 FeS clusters. The first of these, an Fe4S4 cluster, is impressive on its own merit. The second cluster, termed the P cluster, is an outrageous Fe8S7 cluster (one of the biggest naturally occuring FeS cofactors). The uniqueness of nitrogenase does not end there. The next cofactor, an FeMo-co cluster, trumps the lot. It is thought the reaction occurs on the face of this cluster, which incorporates a Molybdenum(!) metal center and a homocitrate molecule just to stabilize the molybdenum (see fig. (b) above).
(2) Electron Transport in Protein: The association of nucleotide-bound Fe-protein subunits to the MoFe protein subunit leads to the release of an electron from ATP towards the first Fe4S4 cluster, which shuttles the electron towards the larger P-cluster. The P-cluster gains further electrons when another Fe-protein binds, eventually passing them on towards the FeMo-co reactive center, which, by the way, has a central carbon atom with 6 bonds to it. Note that this entire process goes on simultaneously on the other half of the protein, making nitrogenase that much more efficient.
(3) Prevention of Oxidation: For a successful dinitrogen reduction to occur, it is imperative that any, and all oxidizing agents are kept well clear of the catalytic site. Rather conveniently, the binding of Fe-proteins subunits to the Fe-Mo protein, using a 'β-grasp' structure, seals off the Fe-Mo protein from oxidation. Even if oxidation were to occur, it would be at the interface of the smaller FeS cluster, thereby protecting the Fe-Mo-co cluster and facilitating further enzymatic reactions to occur.
 |
| Fig. 3: Protein subunit interface. (PDB code: 1N2C. Figure generated using Pymol.) |
Why Nitrogenase Deserves to Win
Simply put, nitrogenase performs the most biologically difficult reactions there is: a reduction of dinitrogen. It so happens, that this reaction is absolutely vital for every living organism. In order to perform this reaction, nitrogenase employs a number of novel structural features, such as two FeS clusters, a molybdenum metal-center, and a carbide, with 6 bonds to carbon. In unison, these unusual features enable nitrogenase to perform some simply phenomenal chemistry.
Should you choose not to vote for it, consider the fate of every protein, DNA & RNA molecule in your body without nitrogen. None of it would polymerize and any bonds that formed would be totally insufficient to support life... all you would be is a pile of oxidized carbon.
References:
- Science , New Series, 2002, 297, 5587, pp. 1696-1700
- Rees, D.C. and Howard, J.B.; Current Opinion in Chemical Biology, 2000, 4, pp. 559-566
- Sgrignani, J.; Franco, D.and Magistrato, A.; Molecules, 2011, 16, pp. 442-465
- Kim, J. and Rees, D.C.; Biochemistry, 1994 33, pp. 389-397
- Hoffman. B.M.; Dean, D.R.; Seefeldt, L.C.; Acc. Chem. Res. 2009, 42, pp. 609-619
- Burgess, B.K. and Lowe, D.J.; Chem. Rev. 1996, 96, pp. 2983-3011
- Evans, D.J. and Pickett, C.J.; Chem. Soc. Rev., 2003, 32, pp. 268–275
- Lancaster. K.M. et al.; Science, 2011: 334 (6058), pp. 974-977
- Goodsell, D.; PDB, February 2002, Molecule of the Month
All protein figures generated using PyMol, from PDB structure 1N2C.