Unraveling the molecular mechanisms of nitrogenase conformational protection against oxygen in diazotrophic bacteria
Nitrogenase serves as the primary protein in the nitrogen
fixation process, converting molecular nitrogen to ammonia. In most cases,
oxygen irreversibly inhibits the function of nitrogenase; however, as the protein
functions in aerobic environments, there is more to the story. This paper
explores the sequence homology of the nitrogenase protein to other
oxygen-sensitive diazotropic bacteria, revealing a method of conformational
protection on the main domain. In addition, surface analysis revealed an
electrostatic surface dimerization domain, where two FeS protein subunits can associate
using a ‘beta-grasp’ mechanism to prevent oxygen damage to the reactive FeS
metallo-cluster. In the process, the protein-coding genes were isolated in A. vinelandii.
 |
| BMC Genomics 2010, 11(Suppl 5):S7 |
Nitrogenase MoFe-Protein at 1.16 Å Resolution: A Central Ligand in theFeMo-Cofactor
Nitrogenase incorporates an Iron-Sulfur cluster
as a cofactor for catalytic activity. The protein performs intra-protein
electron transfer from the homodimeric Fe metalloprotein, via the hydrolysis of
adenosine 5’-triphosphate, to the MoFe protein (an α2β2 tetramer).
It is thought that dinitrogen is reduced in the active site of the FeMo
cofactor, which is examined closely in this study. The authors observed six
central iron atoms which lie on the surface of a sphere with a radius of 2.0 Å
from the cofactor center, coordinated to three inorganic sulfur atoms. These sulfur
atoms of the FeMo-cofactor are themselves equidistant from the center on a
second sphere with a radius of 3.3 Å. The central pocket of this cluster
contains an unidentified atom with a fully occupied valence, closer inspection
identifying it as nitrogen. The geometry enforced by the structure of the
cofactor provides optimal arrangements for dinitrogen reduction.Update: In a recent publication, the central ligand of the FeMoco cofactor was identified as a µ6Carbide...
yes, that is 6 bonds to Carbon! This was identified by Lancaster et. al. using Fe-KEdge spectroscopy and
is reported here.
 |
| Science , New Series, Vol. 297, No. 5587 (Sep. 6, 2002), pp. 1696-1700 |
N2 + 8e- + 16MgATP + 8H+ ⇒ 2NH3 + H2 + 16MgADP + 16Pi (1)
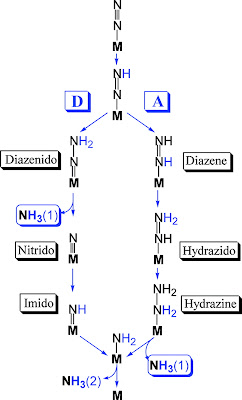
This paper addresses the reaction mechanism and pathway promoted by nitrogenase. Evidently, the 8 electron oxidation is both thermally and kinetically disfavored, leading to the analogy of scaling a peak. The two viable paths to the ‘top’ of the energy barrier are
(a) “distal”: by sequentially adding electrons to each individual nitrogen atom, or
(b) “alternating”: by interchanging between the two atoms of N2.
(a) “distal”: by sequentially adding electrons to each individual nitrogen atom, or
(b) “alternating”: by interchanging between the two atoms of N2.
In addition, the peak analogy persists when considering the difficulty in trapping reaction intermediates to identify substrate-bound species, especially those from the late stages of the reduction pathway. The identification of the reaction pathway is compounded by the mystery of the number, and order, of electrons and protons being delivered to the Mo-Fe cluster during the reaction.
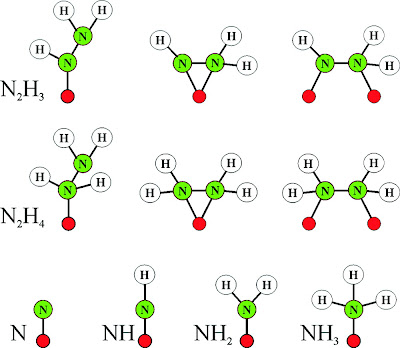 |
| Chart 1. Substrate-Derived Species Bound to FeMo-co That Might Form in Late Stages of N2 Reduction |